R, you may be a confusing and hard to understand language where every package comes with its own set of quirks and foibles. You may make me feel less like a programmer and more like a not-very-well trained magician fumbling around for the right incantation to make magic happen.
But when you work, you do awesome things.
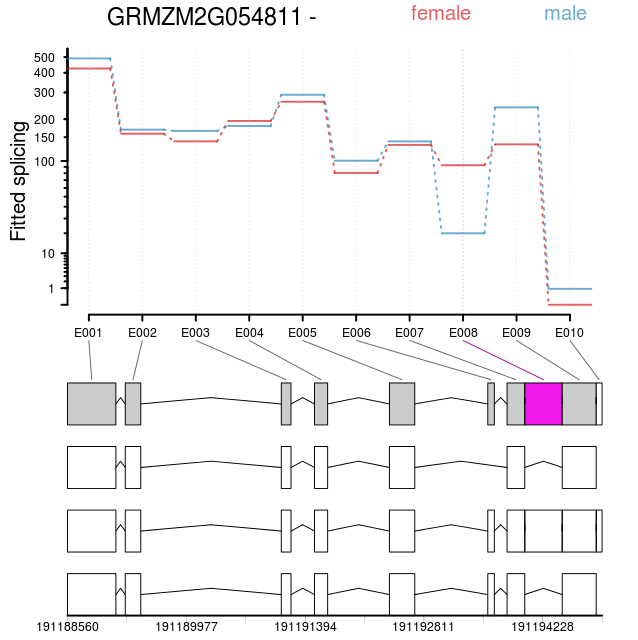
Sex specific splicing of a gene of unknown function of a gene syntenically conserved in all grass species.
With only four days work I was able to go from a giant pile of reads (from the still not properly appreciated Davidson 2011 The Plant Genome) to figures like the one above.
So what is the figure above showing you? One of a large number of genes which show a different pattern of splicing in male and female reproductive organs in maize.* The region “E8” is usually treated as exonic in female reproductive tissues but is spliced out like an intron in male reproductive tissues. What does it mean (if anything)? I have no idea yet! But it would have been a real pain to try to re-invent the wheel for identifying these deferentially spliced genes in python. In R, once I figured out the right incantation, it’s practically plug and play for any gene you could possibly be interested in. Including the software for the (actually quite useful) visualization shown above.
So thank you R. What you do — once I can figure out how to make you do it — you do incredibly well.
*Maize makes it easy for us by separating female and male flowers into two entirely different organs (the ear and tassel respectively).
Data from:
Analyzed using the R package DEXSeq:
Anders S, Reyes A, Huber W. 2012 Detecting differential usage of exons from RNA-Seq Data. Unpublished. (Link is to a PDF)