The corn genome is ~2.4 gigabases (2.4 billion As, Ts, Cs, and Gs) divided among ten chromosomes. The genome of sorghum, the most closely related species with a sequences genome to maize, is also divided into ten chromosomes, but it’s only less than 800 megabases long, approximately a third the size of maize.
What accounts for the size different? Well since their divergence, maize went through a whole genome duplication, doubling it’s genome to twenty chromosomes (which have since been reduced to ten again, as pieces of chromosomes broke apart and stuck to each other*). Since then a bunch of deletions have also occurred, so only sometimes like 20-30% of the genes from the ancestor of maize and sorghum can still be found in both duplicated regions. Clearly the genome duplication of maize is not responsibly (or at least not solely responsible) for the the enormous size of the maize genome.
The real culprit is waves of transposons that have rained across the genome since the whole genome duplication. The maize genome paper estimates that 85% of the genome is composed of transposons. Transposons (“jumping genes”) were discovered first in maize**, although since then they’ve been found in the genomes ranging from bacteria to our own. Transposons are selfish DNA, they replicate as much as they can within the genome of an organism, but don’t*** provide any benefit to their host organism. Every organism has to develop defenses against the replication of transposons or risk being over-run. Some transposons will still move around, which is how they were first discovered in maize, but the majority of kept harmlessly inactive by mechanisms like methylation and RNAi (each of which is at least a whole biology lecture from a trained professor in itself). For some reason, once, or several times in the recent past (recent = last few million years), the transposons in maize have escaped control and run wild, duplicating over and over again, ballooning the genome to its present massive size.
Those same transposons were one of the major hurdles to assembling the maize genome. Is an identical or near identical sequence is present anywhere from hundreds to tens of thousands of times in the genome, figuring out which sequencing reads actually overlap and which just look similar can be really really hard. The longest sequences current technology can generate are only a few thousand base pairs long. Any longer sequence (a single chromosome can be hundreds of millions of base pairs long) must be built up putting together sequences that overlap, like putting together a puzzle.
Given the complexity of the genome, the maize genome assembly is amazingly good, and I stand in awe of the people who put it together. Looking at the pre-release sequence using some tools in our lab, we couldn’t figure out how they’d done such a good job (we were using the sorghum genome as a comparison). With the actual paper out, I can guess some of the “secret sauce” was in the form of incredibly extensive physical, genetic, and optical maps which help line up the fragments in the correct order, but if anything knowing what they had to do to get things to line up so well just makes it more impressive.
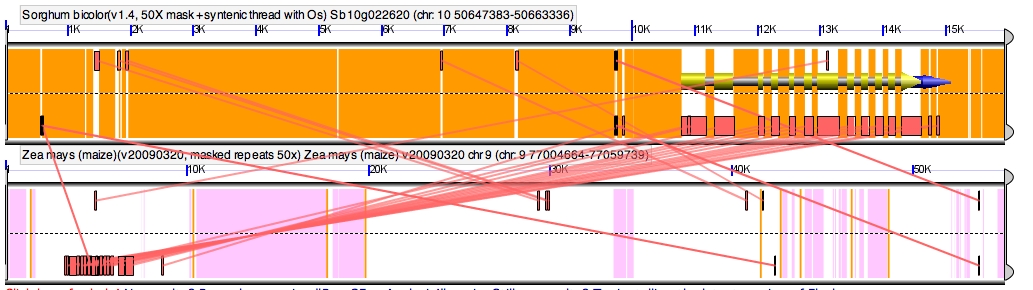
A possible assembly error. Notice that the conserved noncoding sequences of the sorghum gene are backwards relative to the gene with this maize homeolog (boxes on the top represent the DNA read from left to right, and boxes on the bottom reading the inverse sequence right to left). But examples like this are way harder to find than they should be in a the first published release of a genome as complex as maize, and it’s entirely possible this actually represents a cool flipped promoter mutation between maize and sorghum. (The other copy of this gene was also retained after the maize genome duplication, which could compensate if this copy started doing weird/awesome new things instead of fulfilling its ancestral function). You can see this figure yourself in GeVo using this link: http://tinyurl.com/ydupn88
*This process isn’t unique to plants. Humans have 23 chromosomes, our closest relatives, the chimpanzee, has 24. Our second largest chromosome was formed by the merger of two chromosomes from the most recent common ancestor we share with chimps. The two chromosomes (still found seperately in chimps) are creatively names 2A and 2B.
**Transposons were discovered in maize by Barbara McClintock by their effects on the classical genetics of maize traits decades before the structure of DNA was even discovered, let alone a molecular explanation how genes could jump around the genome. She won the Nobel prize for that discovery in 1983. My PI can tell stories about her at Maize Meetings way back in the day, along with the other legends of maize genetics since passed away. I hope someone is keeping a written history of it all.
***Of course some do. Unlike, say, physics, very few rules in biology are without some exceptions.
Trey and I were joking that corn is essentially a mechanism for transposable element delivery 🙂
Comment by Mary — November 20, 2009 @ 1:57 pm
Now that we can answer the question “How Many?” I really want to know the answer to “Why?” especially since the evidence suggests rather than a gradual bloat, most of maize’s transposons originated in one or a couple giant bursts of activity.
Comment by James — November 20, 2009 @ 6:59 pm
Yeah, I know Trey has more qualified thoughts on this as he was a TE researcher in grad school. I was in a different lab so although I heard about his research all the time I wasn’t immersed.
But I remember reading a paper a few months back that has had me thinking. Here’s the GenomeWeb story, Genome Duplication May Have Helped Plants Survive Mass Extinction.
I’ve been wondering about plants shaking up their genomes in times of major stress. You could imagine turning on all your TEs as a crude way to shake up your genome (if you thought like that…). And maybe some fire up better under some stresses than others, or something….?
I’m completely unencumbered by any data on this, and I’m not even caffeinated yet this morning, so maybe that makes no sense. But I wonder if there was a stressor involved?
Comment by Mary — November 21, 2009 @ 8:11 am
It’s definitely a real possibility. I believe there’s been some research which shows that stressing a plant increases the activity of intact transposons. Whether that’s an evolutionary response to increase the chance of new beneficial mutations, or just a case of the plant being too busy to bother repressing all of it’s selfish DNA, it’s a possible explanation.
But then the question would become: What in the last ten million years stressed corn’s teosinte ancestors so much, for so long, that they accumulated 1-2 gigabases of repetitive sequence?
Comment by James — November 21, 2009 @ 11:20 am